♦ Studying the Role of DNA Methylation During Seed Development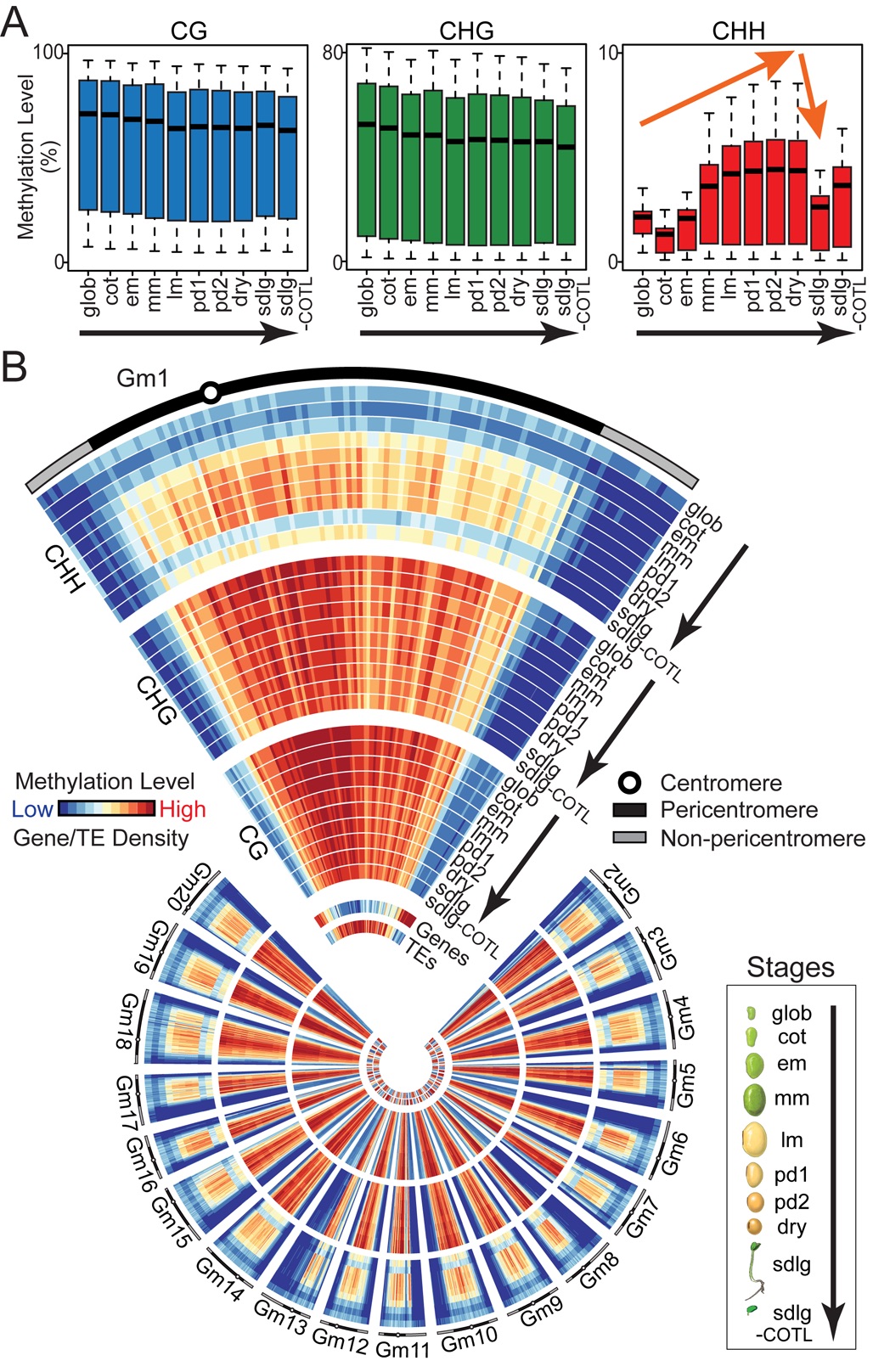
We profiled soybean and Arabidopsis methylomes from the globular stage through dormancy and germination. CHH methylation increases significantly during development throughout the entire seed, targets primarily transposable elements (TEs), is maintained during endoreduplication, and drops precipitously within the germinating seedling. By contrast, no significant global changes in CG- and CHG-context methylation occur during the same developmental period. An Arabidopsis ddcc mutant lacking CHH and CHG methylation does not affect seed development, germination, or major patterns of seed gene expression. These results suggest that CHH and CHG methylation does not play a significant role in seed development, or regulation of seed gene activity - including genes encoding major storage proteins. By contrast, over 100 TEs are de-repressed transcriptionally in ddcc seeds, suggesting that the increase in CHH methylation during seed development may be a failsafe mechanism to reinforce transposon silencing and prevent the occurrence of lethal mutations which may disrupt seed germination.
To date, we have generated 52 BS-Seq Datasets and submitted all to GEO. Click here to download the Bisulfite-Seq datasets.
Publication:
Lin. J., Le, B.H., Min, C., Henry, K.F., Hur, J., Hsieh, T-F., Chen, P-Y., Pelletier, J.M., Pellegrini, M., Fischer, R.L., Harada, J.J., and Goldberg, R.B. (2017). Similarity between soybean and Arabidopsis seed methylomes and loss of non-CG methylation does not affect seed development. Proc. Natl. Acad. Sci. USA. 114, E9730-E9739. [DOWNLOAD PDF]
♦ Seed Genome DNA Methylation Valleys (DMVs) are Enriched For Transcription Factor Genes
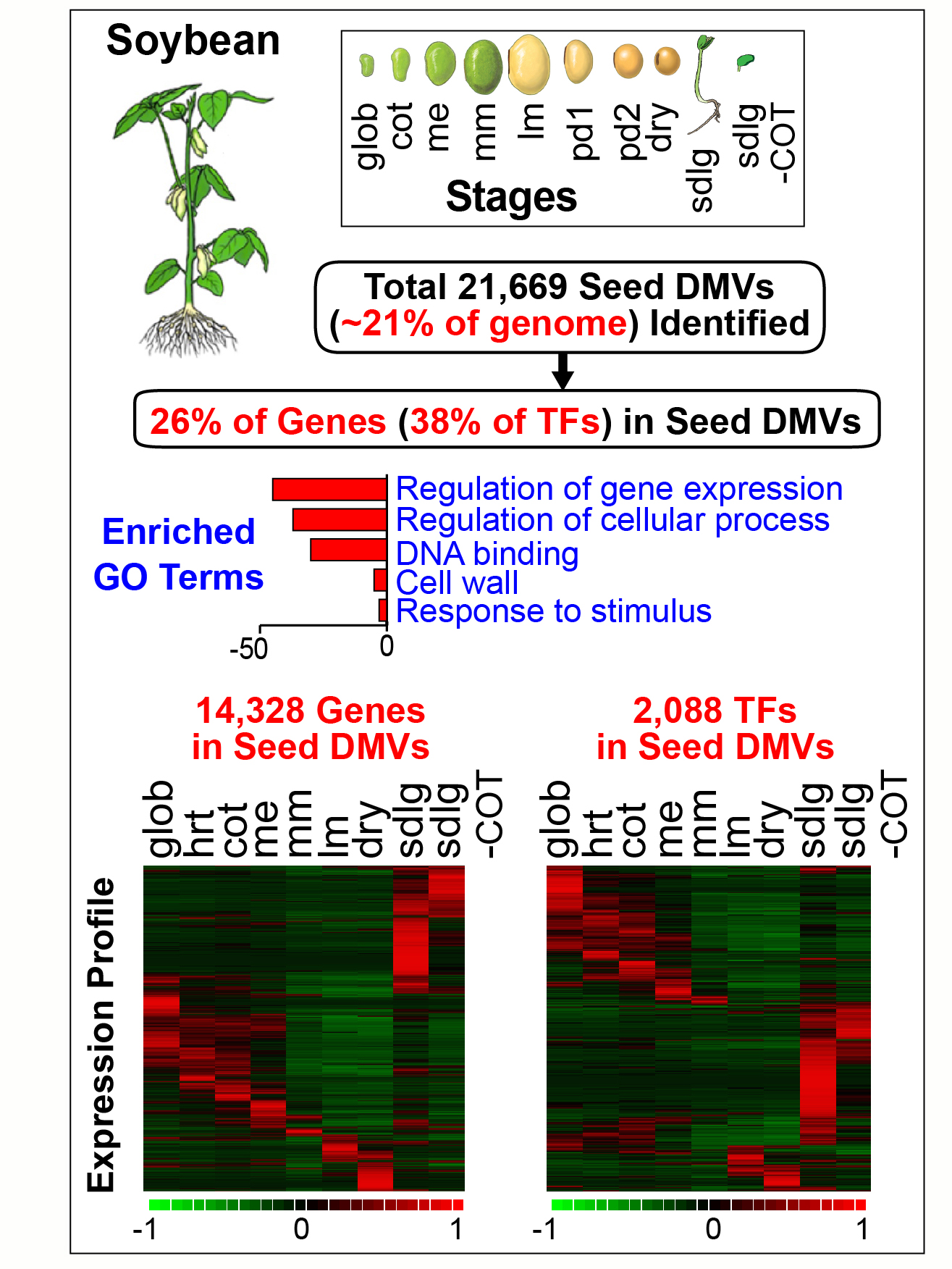
The precise mechanisms that control gene activity during seed development remain largely unknown. In our seed methylome project we showed that several genes essential for seed development-including those encoding storage proteins, fatty acid biosynthesis enzymes, and transcriptional regulators (e.g., ABI3, FUS3) are located within hypomethylated regions of the soybean genome [Lin et al., Proc. Natl. Acad, Sci. USA. 114, E9730-E9739, (2017)]. These hypomethylated regions are similar to the DNA methylation valleys (DMVs), or canyons, found in mammalian cells. In this project we addressed the question of the extent to which DMVs are present within seed genomes and what role they might play in seed development. 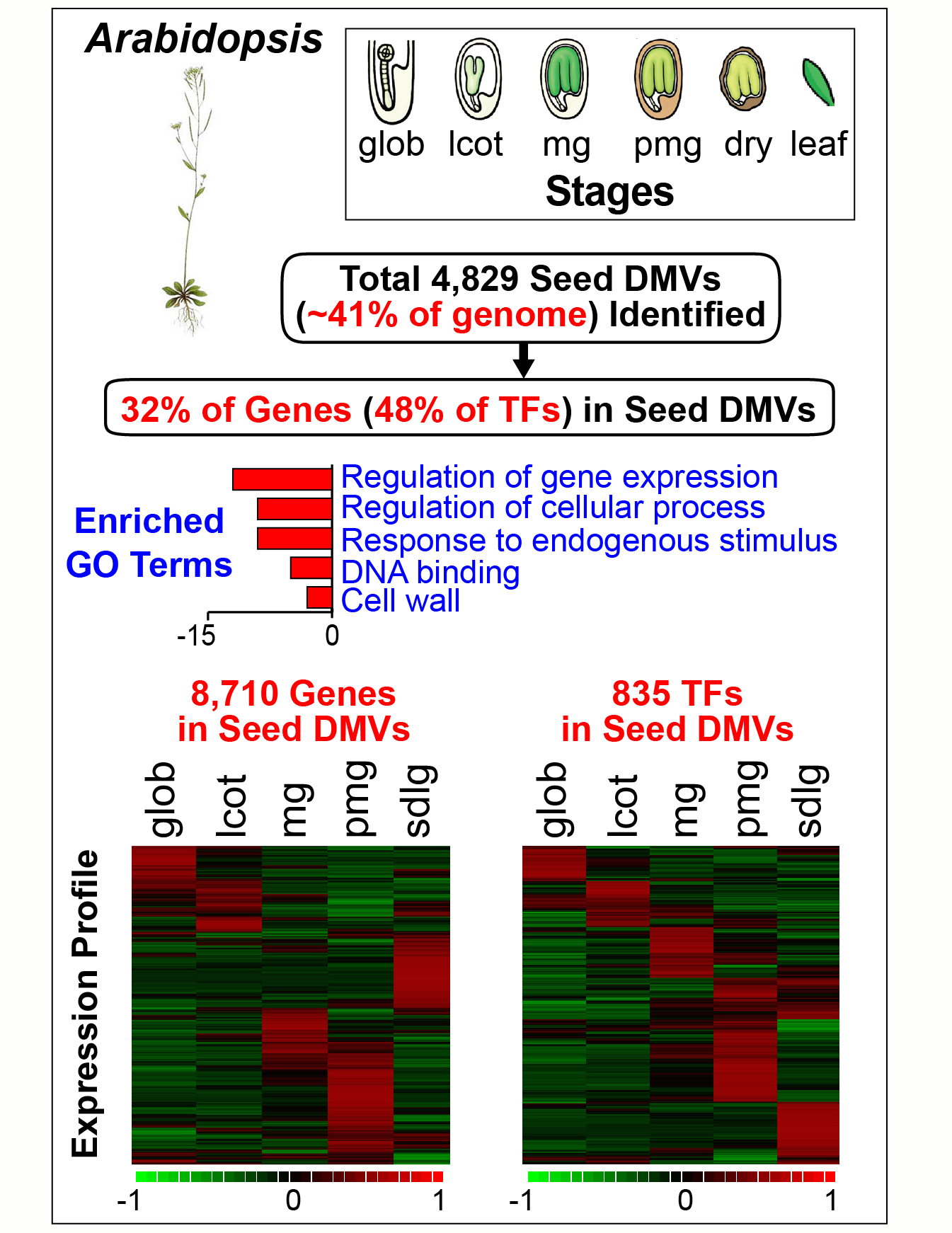 We scanned soybean and Arabidopsis seed genomes from postfertilization through dormancy and germination for regions that contain <5% or <0.4% bulk methylation in CG-, CHG-, and CHH-contexts over all developmental stages. We found that DMVs represent extensive portions of seed genomes, range in size from 5- 76 kb, are scattered throughout all chromosomes, and are hypomethylated throughout the entire plant life cycle. Significantly, DMVs are enriched greatly in transcription factor (TF) genes and other developmental genes that play critical roles in seed formation. Many DMV genes are regulated with respect to seed stage, region, and tissue, and contain H3K4me3, H3K27me3, or bivalent marks that fluctuate during development. Our results indicate that DMVs are a unique regulatory feature of both plant and animal genomes, and that a large number of seed genes are regulated in the absence of methylation changes during development—probably by the action of specific TFs and epigenetic events at the chromatin level.
We scanned soybean and Arabidopsis seed genomes from postfertilization through dormancy and germination for regions that contain <5% or <0.4% bulk methylation in CG-, CHG-, and CHH-contexts over all developmental stages. We found that DMVs represent extensive portions of seed genomes, range in size from 5- 76 kb, are scattered throughout all chromosomes, and are hypomethylated throughout the entire plant life cycle. Significantly, DMVs are enriched greatly in transcription factor (TF) genes and other developmental genes that play critical roles in seed formation. Many DMV genes are regulated with respect to seed stage, region, and tissue, and contain H3K4me3, H3K27me3, or bivalent marks that fluctuate during development. Our results indicate that DMVs are a unique regulatory feature of both plant and animal genomes, and that a large number of seed genes are regulated in the absence of methylation changes during development—probably by the action of specific TFs and epigenetic events at the chromatin level.
Publication:
Chen, M., Lin, J., Hur, J., Pelletier, J.M., Baden, R., Pellegrini, M., Harada, J.J. and Goldberg, R.B. (2018). Seed genome hypomethylated regions are enriched in transcription factor genes. Proc. Natl. Acad. Sci. USA. 115, E8315-E8322.[DOWNLOAD PDF]